Our approaches
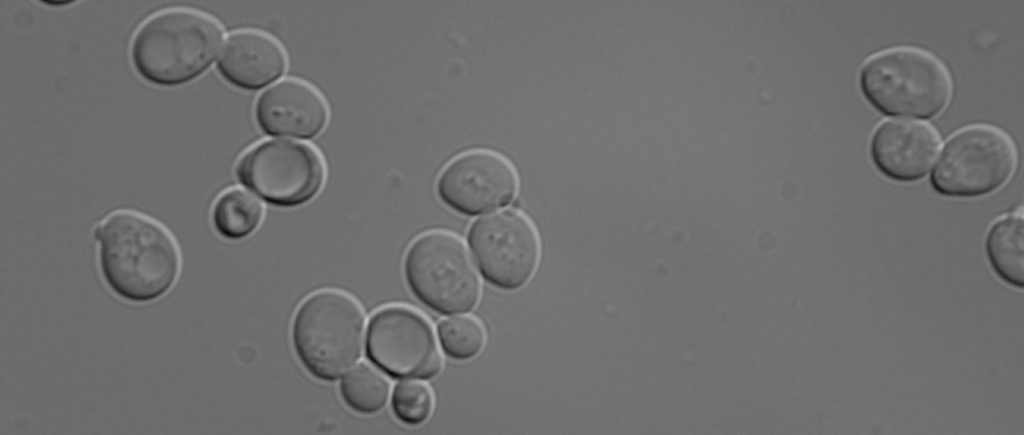
Baker’s Yeast
To unravel how endosomes, autophagosomes, and lysosomes/vacuoles form and function, and how defects in vital proteins cause lysosomal diseases, we use many different approaches.
Our model system is the yeast Saccharomyces cerevisiae, a simple eukaryote. It has not brain, but many highly conserved proteins involved in secretion (Nobel Prize to Randy Schekman, 2013), or autophagy (Nobel Prize to Yoshinori Ohsumi, 2016) were identified in yeast. It is an excellent model system with fast doubling time, excellent genetics and biochemistry. And it smells good!
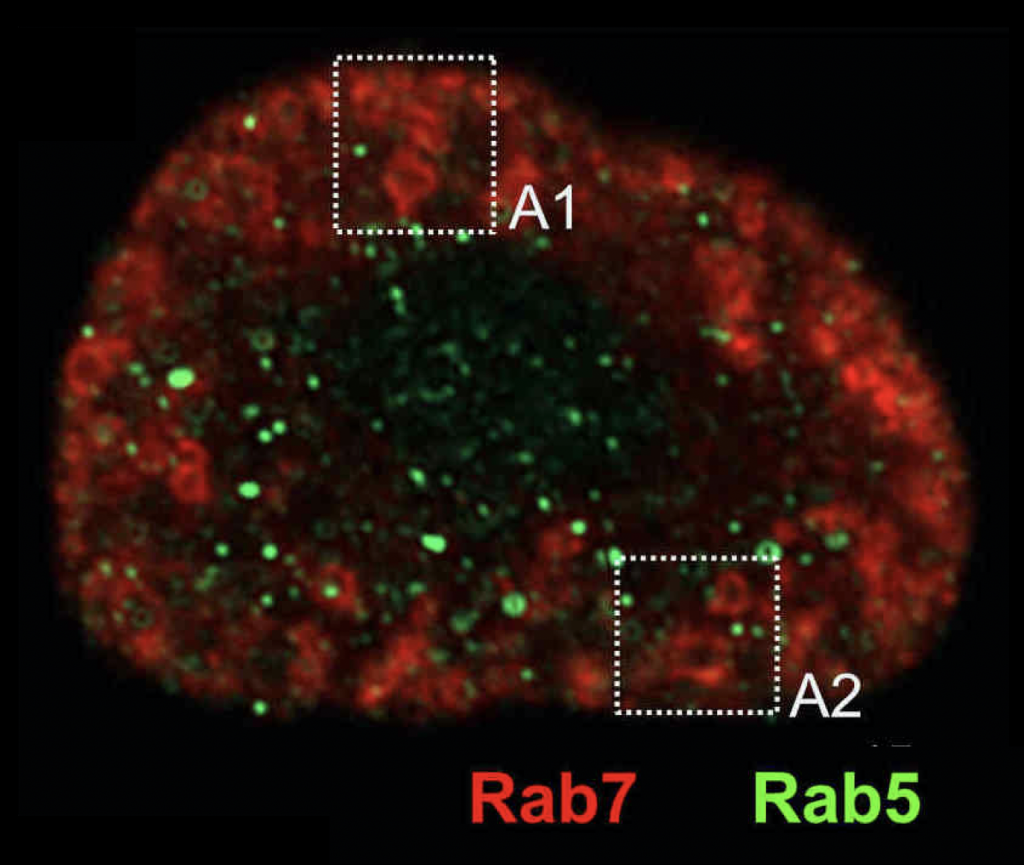
Rabs in Drosophia Nephrocytes
To get insights into the metazoan endosomal and lysosomal system, we also use the fruit fly Drosophila melanogaster, and in particular its kidney-like nephrocytes (with Achim Paululat, Osnabrück).
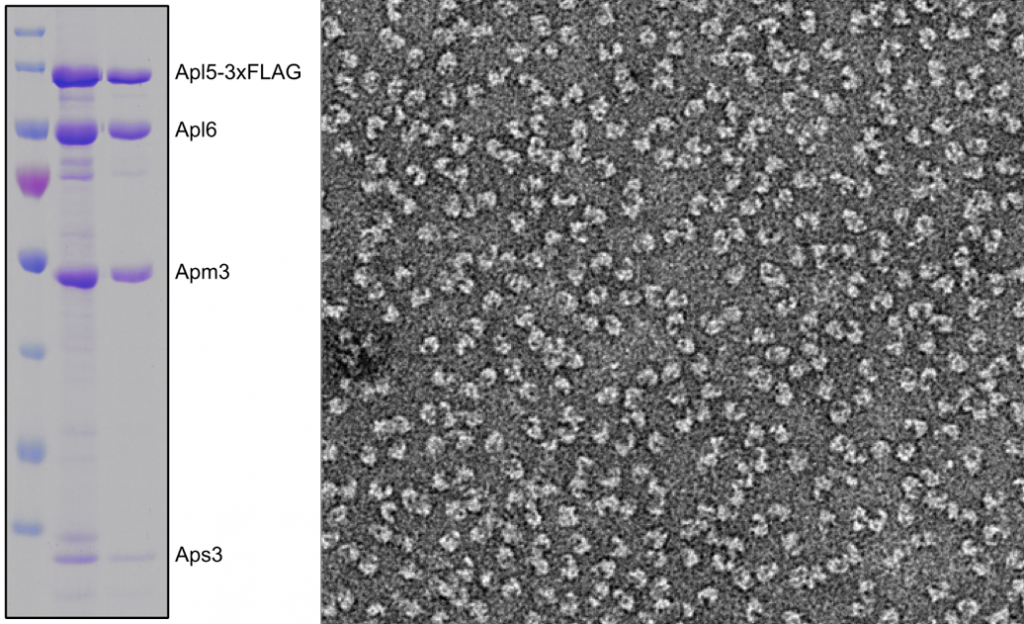
Purified AP-3 complex
Purified Green fluorescent protein
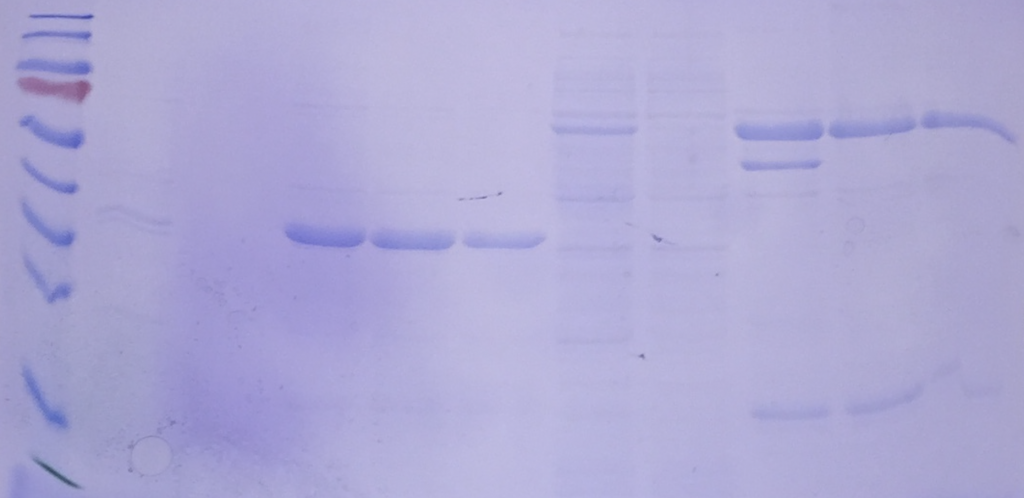
SDS-Gels reveal their purity and size
Biochemistry is the discipline that explains what we call ‚life‘. To ask how proteins function within the endolysosome, we use purified organelles and endosomal or vacuolar proteins and test their function – in Rab activation or inactivation, in membrane tethering or fusion. For many assays, we use liposomes in our in vitro assays. Working with isolated proteins allows us to directly add proteins or just parts of them and test our hypotheses. Biochemistry is for us the gate to a mechanistic understanding of endosomal and lysosomal function. Whereever possible, we work on the structure of proteins in collaborations with colleagues.
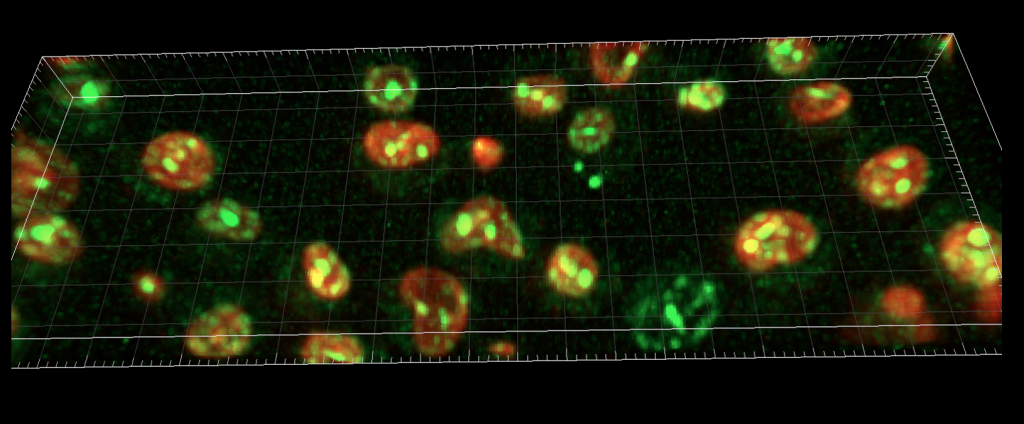
Imaging yeast proteins by lattic light sheet microscopy
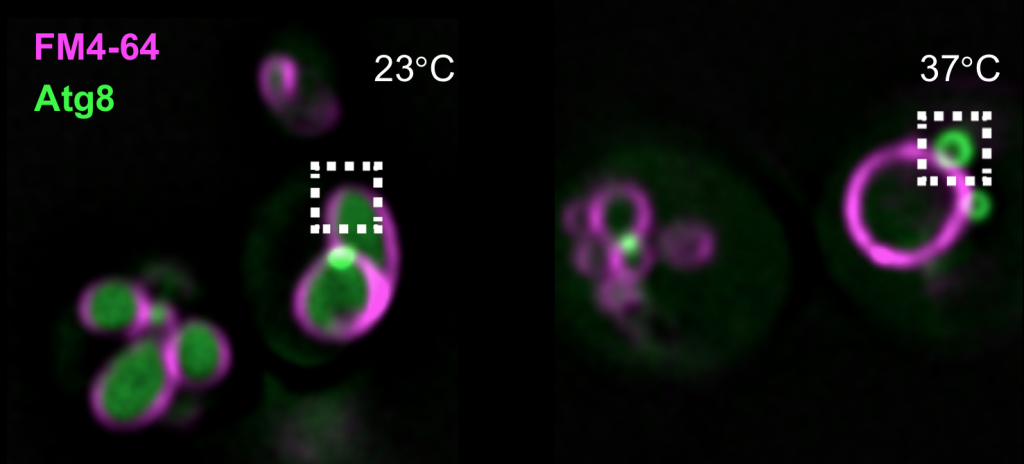
yeast mutants with autophagy defects
Cell Biology approaches – Whenever I ask students, how we gain information on the function of a protein, they suggest to tag it with GFP and then see what it does by microscopy. This is indeed an excellent idea, but imaging per se is useless without further information. Within the lab, we localize proteins relative to each other, analyze how mutations affect their localization and thus learn more about their function.
Mutations in vacuolar proteins can change the shape and size of the vacuole in yeast, and we thus learn about the protein’s function.
In the above examples, we have used the lattice light sheet microscope to look at GFP-tagged proteins at the vacuole by live-cell microscopy (top), and with our Delta Vision microscope at autophagy by following GFP-tagged Atg8.
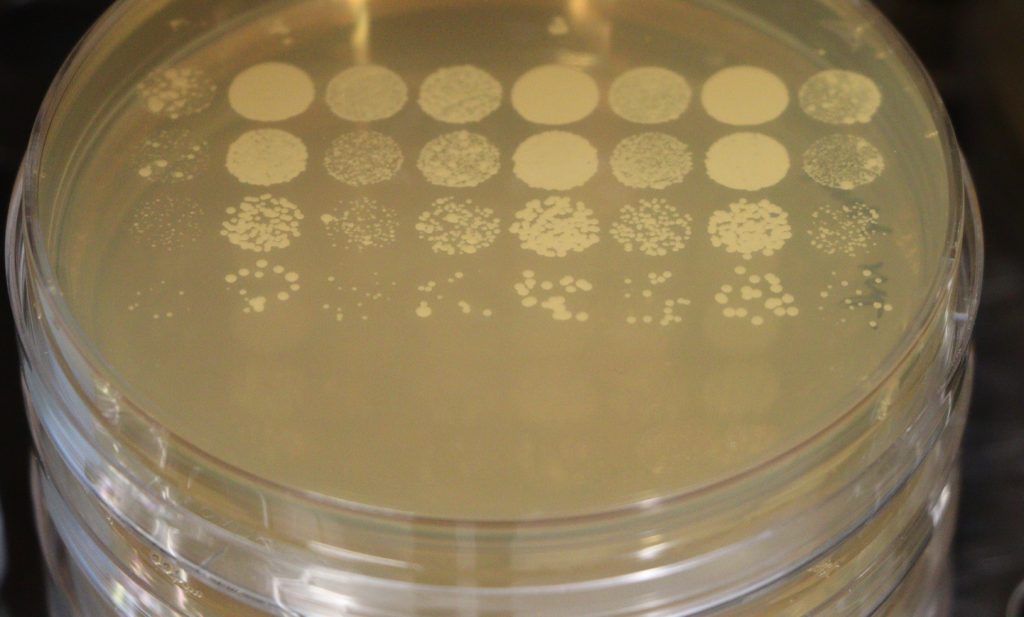
Yeast genetics
Genetics and physiology – Any mutation can cripple a cell, but how do you know about the protein’s function? We use a combination of genetics (we use deletions, mutants generated by homologous recombination or CRISPR/Cas9, complementation etc.), cell biology and biochemistry to uncover the function of endosomal or lysosomal proteins. Until now, we generated some 10,000 yeast mutants in many different experiments as tools to learn how the lysosome functions.
The nice aspect about yeast is that we can let it grow in drops of decreasing cell density and then determine how fast a mutant strain duplicates relative to the wild-type. Growth tells us whether the cell is happy or not – we call this the physiology of the cell.

Center of Cellular Nanoanalytics Osnabrück (CellNanOs)
For all of our experiments, we profit from support by the SFB 1557 collaborative research center, which funds many PhD positions, and the Center of Cellular Nanoanalytics Osnabrück (CellNanOs) with its excellent imaging facilities.
If you would like to know more about our research, see the other pages or contact us!
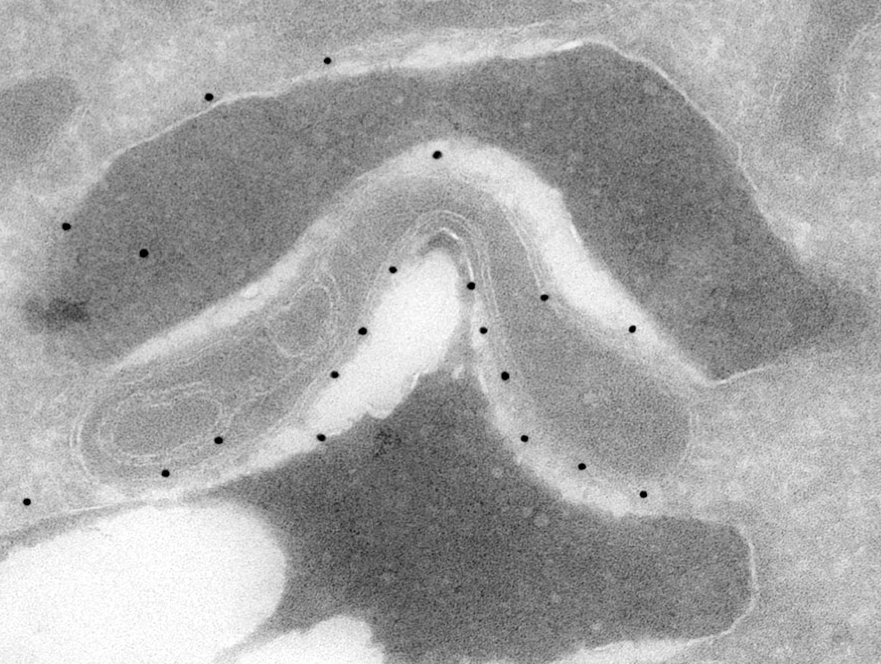
Electron microscopy reveals contact sites of yeast vacuole with mitochondria

